Note
Click here to download the full example code
02: Fitting Power Spectrum Models¶
Introduction to the module, beginning with the FOOOF object.
# Import the FOOOF object
from fooof import FOOOF
# Import utility to download and load example data
from fooof.utils.download import load_fooof_data
# Download examples data files needed for this example
freqs = load_fooof_data('freqs.npy', folder='data')
spectrum = load_fooof_data('spectrum.npy', folder='data')
FOOOF Object¶
At the core of the module, which is object oriented, is the FOOOF object,
which holds relevant data and settings as attributes, and contains methods to run the
algorithm to parameterize neural power spectra.
The organization is similar to sklearn:
A model object is initialized, with relevant settings
The model is used to fit the data
Results can be extracted from the object
Calculating Power Spectra¶
The FOOOF object fits models to power spectra. The module itself does not
compute power spectra, and so computing power spectra needs to be done prior to using
the FOOOF module.
The model is broadly agnostic to exactly how power spectra are computed. Common methods, such as Welch’s method, can be used to compute the spectrum.
If you need a module in Python that has functionality for computing power spectra, try NeuroDSP.
Note that FOOOF objects require frequency and power values passed in as inputs to be in linear spacing. Passing in non-linear spaced data (such logged values) may produce erroneous results.
Fitting an Example Power Spectrum¶
The following example demonstrates fitting a power spectrum model to a single power spectrum.
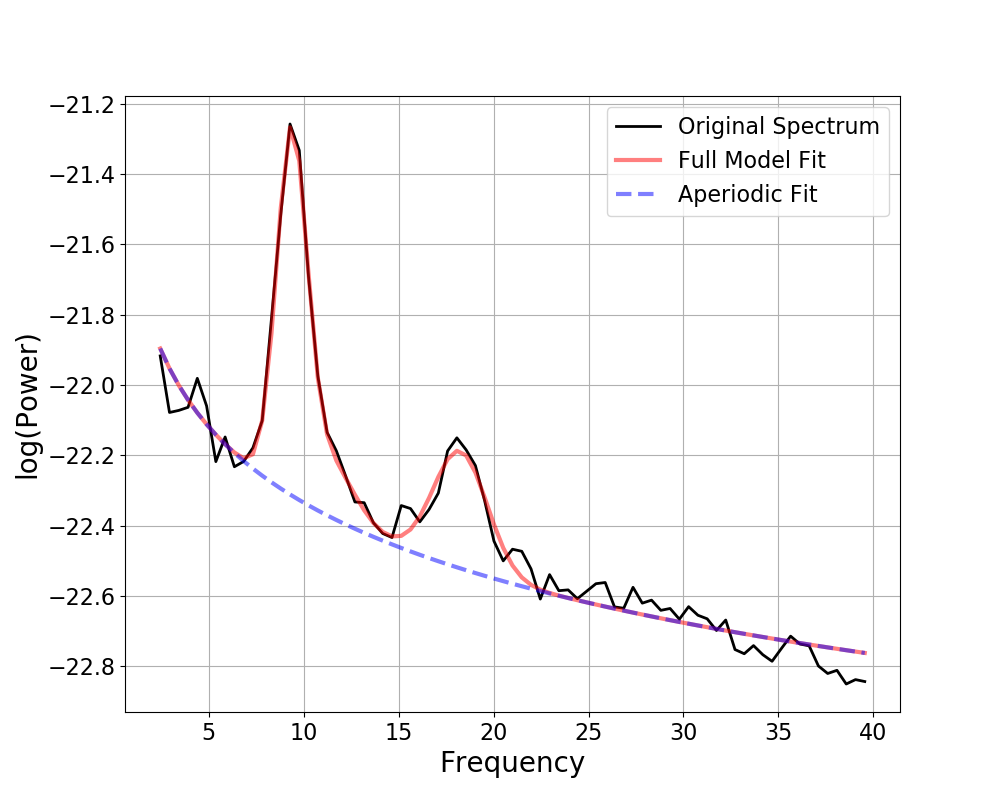
Out:
FOOOF WARNING: Lower-bound peak width limit is < or ~= the frequency resolution: 0.49 <= 0.50
Lower bounds below frequency-resolution have no effect (effective lower bound is the frequency resolution).
Too low a limit may lead to overfitting noise as small bandwidth peaks.
We recommend a lower bound of approximately 2x the frequency resolution.
==================================================================================================
FOOOF - POWER SPECTRUM MODEL
The model was run on the frequency range 2 - 40 Hz
Frequency Resolution is 0.49 Hz
Aperiodic Parameters (offset, exponent):
-21.6185, 0.7160
3 peaks were found:
CF: 9.36, PW: 1.044, BW: 1.59
CF: 11.17, PW: 0.230, BW: 2.88
CF: 18.25, PW: 0.331, BW: 2.85
Goodness of fit metrics:
R^2 of model fit is 0.9829
Error of the fit is 0.0356
==================================================================================================
Fitting Models with ‘Report’¶
The above method ‘report’, is a convenience method that calls a series of methods:
fit(): fits the power spectrum modelprint_results(): prints out the resultsplot(): plots to data and model fit
Each of these methods can also be called individually.
# Alternatively, just fit the model with FOOOF.fit() (without printing anything)
fm.fit(freqs, spectrum, freq_range)
# After fitting, plotting and parameter fitting can be called independently:
# fm.print_results()
# fm.plot()
Out:
FOOOF WARNING: Lower-bound peak width limit is < or ~= the frequency resolution: 0.49 <= 0.50
Lower bounds below frequency-resolution have no effect (effective lower bound is the frequency resolution).
Too low a limit may lead to overfitting noise as small bandwidth peaks.
We recommend a lower bound of approximately 2x the frequency resolution.
Model Parameters¶
Once the power spectrum model has been calculated, the model fit parameters are stored as object attributes that can be accessed after fitting.
Following the sklearn convention, attributes that are fit as a result of the model have a trailing underscore, for example:
aperiodic_params_peak_params_error_r2_n_peaks_
Access model fit parameters from FOOOF object, after fitting:
# Aperiodic parameters
print('Aperiodic parameters: \n', fm.aperiodic_params_, '\n')
# Peak parameters
print('Peak parameters: \n', fm.peak_params_, '\n')
# Goodness of fit measures
print('Goodness of fit:')
print(' Error - ', fm.error_)
print(' R^2 - ', fm.r_squared_, '\n')
# Check how many peaks were fit
print('Number of fit peaks: \n', fm.n_peaks_)
Out:
Aperiodic parameters:
[-21.61849466 0.71602466]
Peak parameters:
[[ 9.36187744 1.04444653 1.58739813]
[11.17320483 0.23009279 2.8755358 ]
[18.24842746 0.33142173 2.84617073]]
Goodness of fit:
Error - 0.035606995591632246
R^2 - 0.9828918513142899
Number of fit peaks:
3
Selecting Parameters¶
You can also select parameters using the get_params()
method, which can be used to specify which parameters you want to extract.
# Extract a model parameter with `get_params`
err = fm.get_params('error')
# Extract parameters, indicating sub-selections of parameter
exp = fm.get_params('aperiodic_params', 'exponent')
cfs = fm.get_params('peak_params', 'CF')
# Print out a custom parameter report
template = ("With an error level of {error:1.2f}, FOOOF fit an exponent "
"of {exponent:1.2f} and peaks of {cfs:s} Hz.")
print(template.format(error=err, exponent=exp,
cfs=' & '.join(map(str, [round(cf, 2) for cf in cfs]))))
Out:
With an error level of 0.04, FOOOF fit an exponent of 0.72 and peaks of 9.36 & 11.17 & 18.25 Hz.
For a full description of how you can access data with get_params(),
check the method’s documentation.
As a reminder, you can access the documentation for a function using ‘?’ in a Jupyter notebook (ex: fm.get_params?), or more generally with the help function in general Python (ex: help(get_params)).
Notes on Interpreting Peak Parameters¶
Peak parameters are labeled as:
CF: center frequency of the extracted peak
PW: power of the peak, over and above the aperiodic component
BW: bandwidth of the extracted peak
Note that the peak parameters that are returned are not exactly the same as the parameters of the Gaussians used internally to fit the peaks.
Specifically:
CF is the exact same as mean parameter of the Gaussian
PW is the height of the model fit above the aperiodic component [1], which is not necessarily the same as the Gaussian height
BW is 2 * the standard deviation of the Gaussian [2]
[1] Since the Gaussians are fit together, if any Gaussians overlap, than the actual height of the fit at a given point can only be assessed when considering all Gaussians. To be better able to interpret heights for single peak fits, we re-define the peak height as above, and label it as ‘power’, as the units of the input data are expected to be units of power.
[2] Gaussian standard deviation is ‘1 sided’, where as the returned BW is ‘2 sided’.
The underlying gaussian parameters are also available from the FOOOF object,
in the gaussian_params_ attribute.
Out:
Peak Parameters Gaussian Parameters
9.36 1.04 1.59 9.36 0.98 0.79
11.17 0.23 2.88 11.17 0.17 1.44
18.25 0.33 2.85 18.25 0.33 1.42
FOOOFResults¶
There is also a convenience method to return all model fit results:
get_results().
This method returns all the model fit parameters, including the underlying Gaussian parameters, collected together into a FOOOFResults object.
The FOOOFResults object, which in Python terms is a named tuple, is a standard data object used with FOOOF to organize and collect parameter data.
# Grab each model fit result with `get_results` to gather all results together
# Note that this returns a FOOOFResult object
fres = fm.get_results()
# You can also unpack all fit parameters when using `get_results`
ap_params, peak_params, r_squared, fit_error, gauss_params = fm.get_results()
# Print out the FOOOFResults
print(fres, '\n')
# From FOOOFResults, you can access the different results
print('Aperiodic Parameters: \n', fres.aperiodic_params)
# Check the r^2 and error of the model fit
print('R-squared: \n {:5.4f}'.format(fm.r_squared_))
print('Fit error: \n {:5.4f}'.format(fm.error_))
Out:
FOOOFResults(aperiodic_params=array([-21.61849466, 0.71602466]), peak_params=array([[ 9.36187744, 1.04444653, 1.58739813],
[11.17320483, 0.23009279, 2.8755358 ],
[18.24842746, 0.33142173, 2.84617073]]), r_squared=0.9828918513142899, error=0.035606995591632246, gaussian_params=array([[ 9.36187744, 0.979158 , 0.79369907],
[11.17320483, 0.16895002, 1.4377679 ],
[18.24842746, 0.33414213, 1.42308536]]))
Aperiodic Parameters:
[-21.61849466 0.71602466]
R-squared:
0.9829
Fit error:
0.0356
Conclusion¶
In this tutorial, we have explored the basics of the FOOOF object,
fitting power spectrum models, and extracting parameters.
Before we move on to controlling the fit procedure, and interpreting the results, in the next tutorial, we will first explore how this model is actually fit.
Total running time of the script: ( 0 minutes 0.554 seconds)