fooof.FOOOFGroup¶
-
class
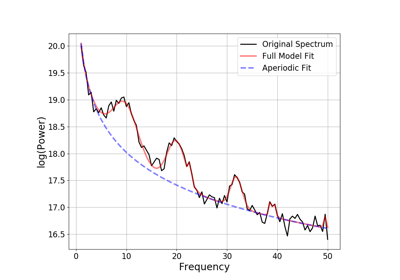
fooof.FOOOFGroup(*args, **kwargs)[source]¶ Model a group of power spectra as a combination of aperiodic and periodic components.
WARNING: FOOOF expects frequency and power values in linear space.
Passing in logged frequencies and/or power spectra is not detected, and will silently produce incorrect results.
- Parameters
- peak_width_limitstuple of (float, float), optional, default: (0.5, 12.0)
Limits on possible peak width, as (lower_bound, upper_bound).
- max_n_peaksint, optional, default: inf
Maximum number of gaussians to be fit in a single spectrum.
- min_peak_heightfloat, optional, default: 0
Absolute threshold for detecting peaks, in units of the input data.
- peak_thresholdfloat, optional, default: 2.0
Relative threshold for detecting peaks, in units of standard deviation of the input data.
- aperiodic_mode{‘fixed’, ‘knee’}
Which approach to take for fitting the aperiodic component.
- verbosebool, optional, default: True
Verbosity mode. If True, prints out warnings and general status updates.
Notes
Commonly used abbreviations used in this module include: CF: center frequency, PW: power, BW: Bandwidth, AP: aperiodic
Input power spectra must be provided in linear scale. Internally they are stored in log10 scale, as this is what the model operates upon.
Input power spectra should be smooth, as overly noisy power spectra may lead to bad fits. For example, raw FFT inputs are not appropriate. Where possible and appropriate, use longer time segments for power spectrum calculation to get smoother power spectra, as this will give better model fits.
The gaussian params are those that define the gaussian of the fit, where as the peak params are a modified version, in which the CF of the peak is the mean of the gaussian, the PW of the peak is the height of the gaussian over and above the aperiodic component, and the BW of the peak, is 2*std of the gaussian (as ‘two sided’ bandwidth).
The FOOOFGroup object inherits from the FOOOF object. As such it also has data attributes (power_spectrum & fooofed_spectrum_), and parameter attributes (aperiodic_params_, peak_params_, gaussian_params_, r_squared_, error_) which are defined in the context of individual model fits. These attributes are used during the fitting process, but in the group context do not store results post-fitting. Rather, all model fit results are collected and stored into the group_results attribute. To access individual parameters of the fit, use the get_params method.
- Attributes
- freqs1d array
Frequency values for the power spectra.
- power_spectra2d array
Power values for the group of power spectra, as [n_power_spectra, n_freqs]. Power values are stored internally in log10 scale.
- freq_rangelist of [float, float]
Frequency range of the power spectra, as [lowest_freq, highest_freq].
- freq_resfloat
Frequency resolution of the power spectra.
- group_resultslist of FOOOFResults
Results of FOOOF model fit for each power spectrum.
has_databoolIndicator for if the object contains data.
has_modelboolIndicator for if the object contains model fits.
n_peaks_intHow many peaks were fit for each model.
- n_failed_fits_int
The number of models that failed to fit.
- failed_fit_inds_list of int
The indices of any models that failed to fit.
Methods
__init__(self, \*args, \*\*kwargs)Initialize object with desired settings.
add_data(self, freqs, power_spectra[, …])Add data (frequencies and power spectrum values) to the current object.
add_meta_data(self, fooof_meta_data)Add data information into object from a FOOOFMetaData object.
add_results(self, fooof_result)Add results data into object from a FOOOFResults object.
add_settings(self, fooof_settings)Add settings into object from a FOOOFSettings object.
copy(self)Return a copy of the current object.
drop(self, inds)Drop one or more model fit results from the object.
fit(self[, freqs, power_spectra, …])Fit a group of power spectra.
get_fooof(self, ind[, regenerate])Get a FOOOF object for a specified model fit.
get_group(self, inds)Get a FOOOFGroup object with the specified sub-selection of model fits.
get_meta_data(self)Return data information from the current object.
get_params(self, name[, col])Return model fit parameters for specified feature(s).
get_results(self)Return the results run across a group of power spectra.
get_settings(self)Return user defined settings of the current object.
load(self, file_name[, file_path])Load FOOOFGroup data from file.
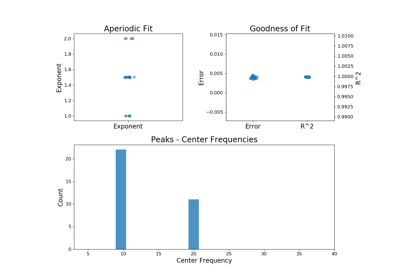
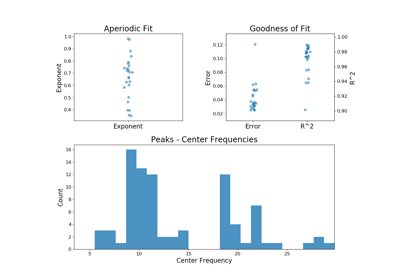
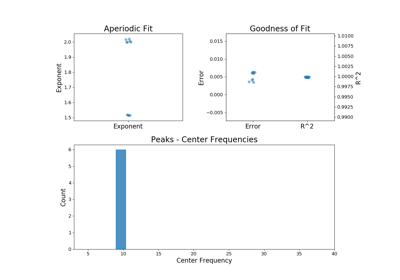
plot(self[, save_fig, file_name, file_path])Plot a figure with subplots visualizing the parameters from a FOOOFGroup object.
print_report_issue([concise])Prints instructions on how to report bugs and/or problematic fits.
print_results(self[, concise])Print out FOOOFGroup results.
print_settings(self[, description, concise])Print out the current settings.
report(self[, freqs, power_spectra, …])Fit a group of power spectra and display a report, with a plot and printed results.
save(self, file_name[, file_path, append, …])Save out results and/or settings from FOOOFGroup object.
save_report(self, file_name[, file_path])Generate and save out a PDF report for a group of power spectrum models.
set_debug_mode(self, debug)Set whether debug mode, wherein an error is raised if fitting is unsuccessful.
Attributes
Indicator for if the object contains data.
Indicator for if the object contains model fits.
How many model fits are null.
How many peaks were fit for each model.
The indices for model fits that are null.
-
add_data(self, freqs, power_spectra, freq_range=None)[source]¶ Add data (frequencies and power spectrum values) to the current object.
- Parameters
- freqs1d array
Frequency values for the power spectra, in linear space.
- power_spectra2d array, shape=[n_power_spectra, n_freqs]
Matrix of power values, in linear space.
- freq_rangelist of [float, float], optional
Frequency range to restrict power spectra to. If not provided, keeps the entire range.
Notes
If called on an object with existing data and/or results these will be cleared by this method call.
-
add_meta_data(self, fooof_meta_data)¶ Add data information into object from a FOOOFMetaData object.
- Parameters
- fooof_meta_dataFOOOFMetaData
A meta data object containing meta data information.
-
add_results(self, fooof_result)¶ Add results data into object from a FOOOFResults object.
- Parameters
- fooof_resultFOOOFResults
A data object containing the results from fitting a FOOOF model.
-
add_settings(self, fooof_settings)¶ Add settings into object from a FOOOFSettings object.
- Parameters
- fooof_settingsFOOOFSettings
A data object containing the settings for a FOOOF model.
-
copy(self)¶ Return a copy of the current object.
-
drop(self, inds)[source]¶ Drop one or more model fit results from the object.
- Parameters
- indsint or array_like of int or array_like of bool
Indices to drop model fit results for. If a boolean mask, True indicates indices to drop.
Notes
This method sets the model fits as null, and preserves the shape of the model fits.
-
fit(self, freqs=None, power_spectra=None, freq_range=None, n_jobs=1, progress=None)[source]¶ Fit a group of power spectra.
- Parameters
- freqs1d array, optional
Frequency values for the power_spectra, in linear space.
- power_spectra2d array, shape: [n_power_spectra, n_freqs], optional
Matrix of power spectrum values, in linear space.
- freq_rangelist of [float, float], optional
Desired frequency range to run FOOOF on. If not provided, fits the entire given range.
- n_jobsint, optional, default: 1
Number of jobs to run in parallel. 1 is no parallelization. -1 uses all available cores.
- progress{None, ‘tqdm’, ‘tqdm.notebook’}, optional
Which kind of progress bar to use. If None, no progress bar is used.
Notes
Data is optional, if data has already been added to the object.
-
get_fooof(self, ind, regenerate=True)[source]¶ Get a FOOOF object for a specified model fit.
- Parameters
- indint
The index of the FOOOFResults in FOOOFGroup.group_results to load.
- regeneratebool, optional, default: False
Whether to regenerate the model fits from the given fit parameters.
- Returns
- fmFOOOF
The FOOOFResults data loaded into a FOOOF object.
-
get_group(self, inds)[source]¶ Get a FOOOFGroup object with the specified sub-selection of model fits.
- Parameters
- indsarray_like of int or array_like of bool
Indices to extract from the object. If a boolean mask, True indicates indices to select.
- Returns
- fgFOOOFGroup
The requested selection of results data loaded into a new FOOOFGroup object.
-
get_meta_data(self)¶ Return data information from the current object.
- Returns
- FOOOFMetaData
Object containing meta data from the current object.
-
get_params(self, name, col=None)[source]¶ Return model fit parameters for specified feature(s).
- Parameters
- name{‘aperiodic_params’, ‘peak_params’, ‘gaussian_params’, ‘error’, ‘r_squared’}
Name of the data field to extract across the group.
- col{‘CF’, ‘PW’, ‘BW’, ‘offset’, ‘knee’, ‘exponent’} or int, optional
Column name / index to extract from selected data, if requested. Only used for name of {‘aperiodic_params’, ‘peak_params’, ‘gaussian_params’}.
- Returns
- outndarray
Requested data.
- Raises
- NoModelError
If there are no model fit results available.
- ValueError
If the input for the col input is not understood.
Notes
For further description of the data you can extract, check the FOOOFResults documentation.
-
get_settings(self)¶ Return user defined settings of the current object.
- Returns
- FOOOFSettings
Object containing the settings from the current object.
-
has_data¶ Indicator for if the object contains data.
-
has_model¶ Indicator for if the object contains model fits.
-
load(self, file_name, file_path=None)[source]¶ Load FOOOFGroup data from file.
- Parameters
- file_namestr
File to load data from.
- file_pathstr, optional
Path to directory to load from. If None, loads from current directory.
-
n_null_¶ How many model fits are null.
-
n_peaks_¶ How many peaks were fit for each model.
-
null_inds_¶ The indices for model fits that are null.
-
plot(self, save_fig=False, file_name=None, file_path=None)[source]¶ Plot a figure with subplots visualizing the parameters from a FOOOFGroup object.
- Parameters
- save_figbool, optional, default: False
Whether to save out a copy of the plot.
- file_namestr, optional
Name to give the saved out file.
- file_pathstr, optional
Path to directory to save to. If None, saves to current directory.
- Raises
- NoModelError
If the FOOOF object does not have model fit data available to plot.
-
static
print_report_issue(concise=False)¶ Prints instructions on how to report bugs and/or problematic fits.
- Parameters
- concisebool, optional, default: False
Whether to print the report in a concise mode, or not.
-
print_results(self, concise=False)[source]¶ Print out FOOOFGroup results.
- Parameters
- concisebool, optional, default: False
Whether to print the report in a concise mode, or not.
-
print_settings(self, description=False, concise=False)¶ Print out the current settings.
- Parameters
- descriptionbool, optional, default: False
Whether to print out a description with current settings.
- concisebool, optional, default: False
Whether to print the report in a concise mode, or not.
-
report(self, freqs=None, power_spectra=None, freq_range=None, n_jobs=1, progress=None)[source]¶ Fit a group of power spectra and display a report, with a plot and printed results.
- Parameters
- freqs1d array, optional
Frequency values for the power_spectra, in linear space.
- power_spectra2d array, shape: [n_power_spectra, n_freqs], optional
Matrix of power spectrum values, in linear space.
- freq_rangelist of [float, float], optional
Desired frequency range to run FOOOF on. If not provided, fits the entire given range.
- n_jobsint, optional, default: 1
Number of jobs to run in parallel. 1 is no parallelization. -1 uses all available cores.
- progress{None, ‘tqdm’, ‘tqdm.notebook’}, optional
Which kind of progress bar to use. If None, no progress bar is used.
Notes
Data is optional, if data has already been added to the object.
-
save(self, file_name, file_path=None, append=False, save_results=False, save_settings=False, save_data=False)[source]¶ Save out results and/or settings from FOOOFGroup object. Saves out to a JSON file.
- Parameters
- file_namestr or FileObject
File to save data to.
- file_pathstr, optional
Path to directory to load from. If None, loads from current directory.
- appendbool, optional, default: False
Whether to append to an existing file, if available. This option is only valid (and only used) if ‘file_name’ is a str.
- save_resultsbool, optional
Whether to save out FOOOF model fit results.
- save_settingsbool, optional
Whether to save out FOOOF settings.
- save_databool, optional
Whether to save out power spectra data.
- Raises
- ValueError
If the data or save file specified are not understood.
-
save_report(self, file_name, file_path=None)[source]¶ Generate and save out a PDF report for a group of power spectrum models.
- Parameters
- file_namestr
Name to give the saved out file.
- file_pathstr, optional
Path to directory to save to. If None, saves to current directory.
-
set_debug_mode(self, debug)¶ Set whether debug mode, wherein an error is raised if fitting is unsuccessful.
- Parameters
- debugbool
Whether to run in debug mode.